NGS
-
Application note: A comprehensive method protocol for annotation and integrated functional understanding of lncRNAs
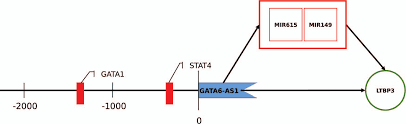 Long non-coding RNAs (lncRNAs) are of fundamental biological importance; however, their functional role is often unclear or loosely defined as experimental characterization is challenging and bioinformatic methods are limited. We developed a novel integrated method protocol for the annotation and detailed functional characterization of lncRNAs within the genome. It combines annotation, normalization and gene expression with sequence-structure conservation, functional interactome and promoter analysis. Our protocol allows an analysis based on the tissue and biological context, and is powerful in functional characterization of experimental and clinical RNA-Seq datasets including existing lncRNAs. This is demonstrated on the uncharacterized lncRNA GATA6-AS1 in dilated cardiomyopathy.
Long non-coding RNAs (lncRNAs) are of fundamental biological importance; however, their functional role is often unclear or loosely defined as experimental characterization is challenging and bioinformatic methods are limited. We developed a novel integrated method protocol for the annotation and detailed functional characterization of lncRNAs within the genome. It combines annotation, normalization and gene expression with sequence-structure conservation, functional interactome and promoter analysis. Our protocol allows an analysis based on the tissue and biological context, and is powerful in functional characterization of experimental and clinical RNA-Seq datasets including existing lncRNAs. This is demonstrated on the uncharacterized lncRNA GATA6-AS1 in dilated cardiomyopathy.You can find here the supplement information describing in detail the workflow combining gensearchNGS with other tools used in the work published here:
A comprehensive method protocol for annotation and integrated functional understanding of lncRNAs
Meik Kunz, Beat Wolf, Maximilian Fuchs, Jan Christoph, Ke Xiao, Thomas Thum, David Atlan, Hans-Ulrich Prokosch, Thomas Dandekar
Briefings in Bioinformatics, Volume 21, Issue 4, July 2020, Pages 1391–1396, https://doi.org/10.1093/bib/bbz066
Published: 03 October 2019 -
Application note: Homozygous Inversion on Chromosome 13 Involving SGCG Detected by Short Read Whole Genome Sequencing in a Patient Suffering from Limb-Girdle Muscular Dystrophy
This paper describes a method to detect a long (around 350kb) homozygous inversion by WGS using GensearchNGS.
You can find the work published here:
Homozygous Inversion on Chromosome 13 Involving SGCG Detected by Short Read Whole Genome Sequencing in a Patient Suffering from Limb-Girdle Muscular Dystrophy
Authors: Natalie Pluta, Sabine Hoffjan, Frederic Zimmer, Cornelia Köhler, Thomas Lücke, Jennifer Mohr, Matthias Vorgerd, Hoa Huu Phuc Nguyen, David Atlan, Beat Wolf, Ann-Kathrin Zaum and Simone Rost
Publication: Genes 2022, 13(10), 1752; https://doi.org/10.3390/genes13101752 -
Clinvar introduces submission API
Clinvar has introduced an API to submit variant data. We will implement this shortly into our Gensearch and GensearchNGS products to replace the old FTP style semi-automatic submission tool.
In the meantime, please check whether your support and maintenance contract is up to date, if not, please contact your sales representative to extend your contract.
-
Wuhan COVID19 flu
 As the coronavirus disease hits pandemic proportions we have included the current ASM985889v3 RefSeq to help support the research community in the field as well as technical improvements to our software to ease user inclusion of updates of this RefSeq that will certainly be deposited in the NCBI databases. Phenosystems SA has also committed to provide free of charge one year licenses to any laboratory, private or public, wishing to analyse SARS-CoV2 or other related coronaviruses in this fight against COVID-19. We will also provide support to these labs as far as possible during these difficult times.
As the coronavirus disease hits pandemic proportions we have included the current ASM985889v3 RefSeq to help support the research community in the field as well as technical improvements to our software to ease user inclusion of updates of this RefSeq that will certainly be deposited in the NCBI databases. Phenosystems SA has also committed to provide free of charge one year licenses to any laboratory, private or public, wishing to analyse SARS-CoV2 or other related coronaviruses in this fight against COVID-19. We will also provide support to these labs as far as possible during these difficult times.You can download the application note here.
Please contact us for further information and the free licences.
-
Wuhan COVID19 upload module for German RKI
 Germany has launched a national effort to fund sequencing of up to 10% of PCR samples taken from patients. A central database is being build at the Robert Koch Institut tasked to collect those sequences.
Germany has launched a national effort to fund sequencing of up to 10% of PCR samples taken from patients. A central database is being build at the Robert Koch Institut tasked to collect those sequences.We have implemented a first version of an export module compatible with the requirements defined by the Robert Koch Institut.
You can download the latest version of our application note here.
This email address is being protected from spambots. You need JavaScript enabled to view it.